1. Distribution model#
Import the necessary functionalities from Relife
[1]:
import numpy as np
import matplotlib.pyplot as plt
from relife import Weibull, Gompertz
from relife.datasets import load_circuit_breaker
Here is a toy datasets that contains the following 15 first data
[2]:
import pandas as pd
time, event, entry = load_circuit_breaker()
data = pd.DataFrame({"time": time, "event": event, "entry": entry})
display(data.head(n=15))
| time | event | entry | |
|---|---|---|---|
| 0 | 34.0 | 1.0 | 33.0 |
| 1 | 28.0 | 1.0 | 27.0 |
| 2 | 12.0 | 1.0 | 11.0 |
| 3 | 38.0 | 1.0 | 37.0 |
| 4 | 18.0 | 1.0 | 17.0 |
| 5 | 32.0 | 1.0 | 31.0 |
| 6 | 44.0 | 1.0 | 43.0 |
| 7 | 49.0 | 1.0 | 48.0 |
| 8 | 27.0 | 1.0 | 26.0 |
| 9 | 47.0 | 1.0 | 46.0 |
| 10 | 44.0 | 1.0 | 43.0 |
| 11 | 70.0 | 1.0 | 69.0 |
| 12 | 40.0 | 1.0 | 38.0 |
| 13 | 37.0 | 1.0 | 35.0 |
| 14 | 26.0 | 1.0 | 24.0 |
1.1. Basics#
One can instanciate a Weibull distribution model as follow
[3]:
weibull = Weibull()
From now, the model parameters are unknow, thus set to np.nan
[4]:
print(weibull.params_names)
print(weibull.params)
['shape', 'rate']
[None None]
1.2. Parameters estimations#
One can fit the model. You can either return a new fitted instance or fit the model inplace
[5]:
weibull.fit(time, event, entry, inplace=True)
print("Estimated parameters are :", weibull.params)
Estimated parameters are : [3.7267452 0.01232326]
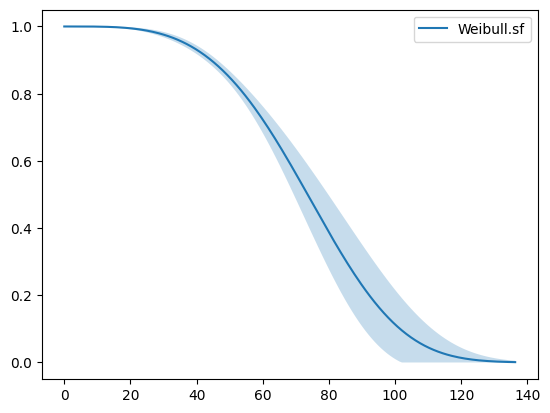
1.3. Plots#
To plot the survival function, do the following
[6]:
weibull.plot.sf()
[6]:
<Axes: >